Fluorescence in situ hybridization (FISH) vs Genomic in situ hybridization (GISH)
In situ hybridization techniques, such as fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH), is widely used to identify chromosome morphologies and sequences, amount and distribution of various types of chromatin in chromosomes, and genome organization during the metaphase stage of meiosis.

Fluorescence in situ hybridization (FISH) is a laboratory technique for detecting and locating a specific DNA sequence or a gene on a chromosome within a person’s genome.
The technique relies on exposing chromosomes to a small DNA sequence called a probe that has a fluorescent molecule attached to it.
FISH helps scientist to visualize the location of particular gene to check for a variety of chromosomal abnormalities.

Summary of steps
1. Cells cultured, harvested, prepared on microscopic slides and are denatured (now DNA is single stranded for probe attachment)
Cells on metaphase stage of division is selected (as maximum condensation on metaphase stage)
2. Fluorescently labeled hybridization probe is added
(The hybridization probe is a short fragment of DNA that has a fluorescent dye attached that enable scientist to visualize the site of probe attachment. A typical FISH probe would be 10 - 100 kb long)
3. If the DNA corresponding to the probe is present in the sample, then the fluorescently labeled probe will attach to the DNA and will be visible under a fluorescent microscope.
4. This allows deletions (no fluorescent spot at the expected position) and rearrangements (spot present, but in an unexpected chromosomal location) to be detected. Thus helps in diagnosis of genetic diseases.

Figure. An example of FISH-treated metaphase chromosomes: Here, chromosomes 1, 2, and 4 were labeled yellow with FISH and the other chromosomes were stained red. Translocations between yellow and red chromosomes are detected. The left picture represents a normal cell (the numbers in the figure indicate chromosome numbers) and the right picture is an example of reciprocal translocation with two bi-color chromosomes (indicated by two arrows). http://www.rerf.jp/dept/genetics/fish_e.html
Applications:- To detect chromosomal aberrations or abnormalities in humans
- To study somaclonal variation* in plants
- In chromosome mapping
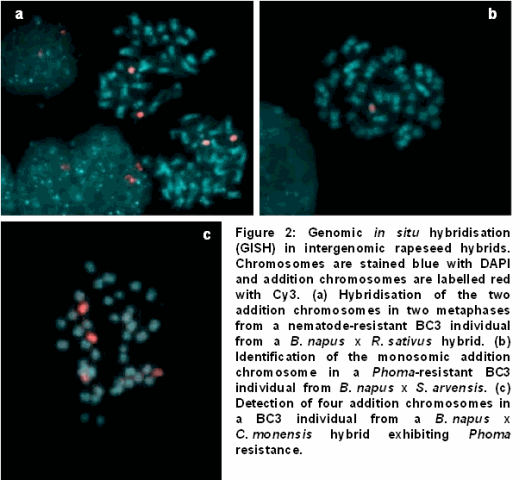
Summary of Steps
- Extraction of total genomic DNA of one of the species of interest (to be used as probe)
- Chromosome preparations of the species 2 being studied
- Repeated sequences in both species anneal quickly than the unique sequences of the genome
- Thus helps in assessing genome relationship between species
- GISH allows characterization of the genome and chromosome of hybrid plats and recombinant breeding lines
- Helps in assessing phylogenetic relationship between different species of plants
· Devi, J., Ko, J. M., & Seo, B. B. (2005). FISH and GISH: Modern cytogenetic techniques. Indian Journal of Biotechnology, 4(3), 307.
· Younis, A., Ramzan, F., Hwang, Y. J., & Lim, K. B. (2015). FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Reports, 34(9), 1477-1488.
· http://www.geneticseducation.nhs.uk/laboratory-process-and-testing-techniques/fish
· Snowdon, R. J., Köhler, A., Köhler, W., & Friedt, W. (1999). FISHing for new rapeseed lines: the application of molecular cytogenetic techniques to Brassica breeding. New horizons for an old crop. Proc 10th Int Rapeseed Congr. The Regional Institute, Gosford, NSW, Australia. (GISH image credit)
*Somaclonal variation: genetic variation in plants raised by tissue culture.
Really its a amazing post.... Short and understandable..... Loved this 😊
ReplyDeletenicely presented
ReplyDeleteShort n easily understable.
ReplyDeleteNice presentation
Nice
ReplyDeletewell understable post tnks.
ReplyDeletebt i wnt sm more topics on ph.d entrens level.
what use for this techniques
ReplyDeletewell defined and thank you for enriching our knowledge...….
ReplyDeletePost a Comment
We Love to hear from U :) Leave us a Comment to improve this site
Thanks for Visiting.....